When a medical Vision Language Model(VLM) looks at a chest X-ray and says “cardiomegaly present,” what’s actually happening inside the model? It’s a black box. Billions of parameters. Dense activation vectors where every dimension encodes a tangled mix of concepts. This is my experimentation using Sparse Autoencoders (SAEs) to interpret MedGemma, Google’s vision-language model for medical imaging. Thanks to DeepMind Team for an open release of Gemma Scope
BlackBox : Neural Networks Are Opaque
MedGemma has a hidden dimension of 2,560. When you feed it a chest X-ray and ask about cardiomegaly, the model produces an activation vector that looks something like this:
[0.23, -0.15, 0.42, 0.08, -0.31, ...]
Every single one of those 2,560 numbers represents a mixture of many concepts. This is called superposition. The model packs more ideas than it has dimensions by encoding them as overlapping patterns. It is next to impossible to figuring out what any single value means.
SAEs provide a Way In
SAEs learn to decompose those dense vectors into a much larger set of sparse features. Instead of 2,560 tangled dimensions, we get 65,000+ features where:
Each feature tends to represent a single concept
Most features are zero for any given input
We can focus on the handful that actually matter
My mental model for this goes like this, dense vector is like hearing an entire orchestra playing at once. The SAE separates out each instrument so you can listen to the violin, the cello, the flute individually.
Dense activation → SAE Encoder → Sparse features (65k dims)
[0.23, -0.15, ...] → [0, 0, 0, 142, 0, 0, 89, 0, 0, ...]
↑ ↑
Feature 3818 Feature 7241
"formal tone" "lung region"
Using GemmaScope 2 on MedGemma
Training SAEs from scratch takes serious compute. Fortunately, Google released GemmaScope 2, a suite of pre-trained SAEs for the Gemma model family. One caveat is that GemmaScope 2 was trained on general Gemma 3 activations. MedGemma was fine-tuned for medical tasks. Would the SAE even work?
Based on my analysis, it appears so.I measured reconstruction quality using Fraction of Variance Unexplained (FVU). Lower is better. Here’s what I found across different layers:
| Layer | FVU | Variance Explained |
|---|---|---|
| 9 | 0.006 | 99.4% |
| 17 | 0.020 | 98.0% |
| 22 | 0.013 | 98.7% |
| 29 | 0.053 | 94.7% |
Medical fine-tuning didn’t dramatically shift the activation distributions. The SAE captures over 95% of variance across all layers. That’s enough for interpretability work.A few caveats though. Later layers show more drift. And the feature meanings might shift: a “formal language” feature in general Gemma 3 might fire on clinical terminology in MedGemma.
Experiment 1: Different Questions, Different Circuits
I loaded a sample chest X-ray and asked MedGemma four different clinical questions:
Is there cardiomegaly?
Is there pneumonia?
Is there a pleural effusion?
Is this a normal chest X-ray?
Same image. Different questions. What happened in the feature space?
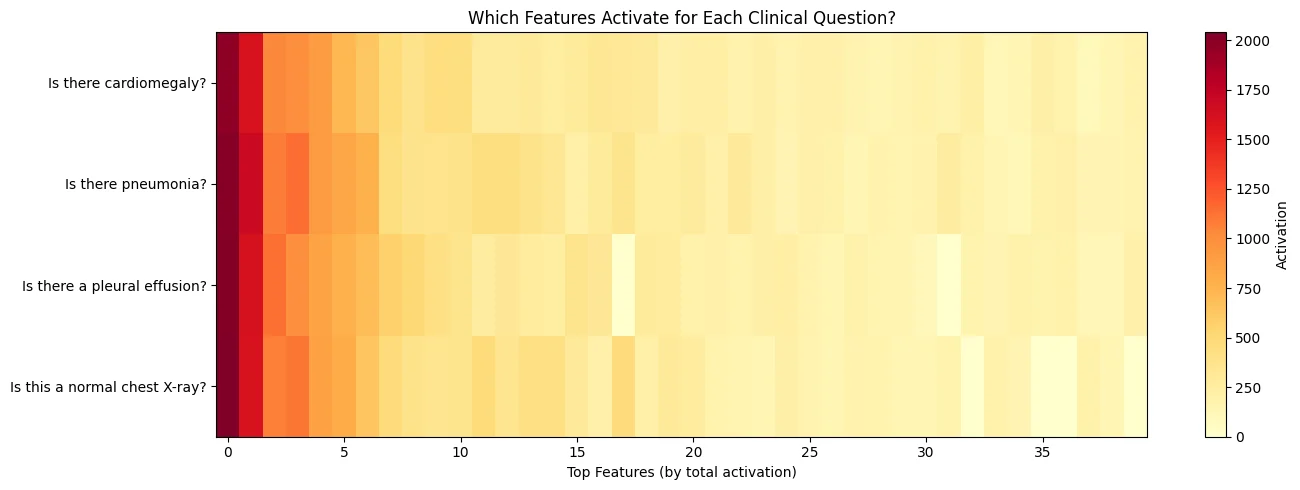
Each question activated a distinct pattern of features. The “cardiomegaly” question lit up 77 features. The “pleural effusion” question lit up 90. The overlap was substantial (cosine similarity above 0.9), but each pathology had its own signature. I think this makes sense. The model is routing different clinical concepts through different internal circuits.
Experiment 2: The Phrasing Effect
Here’s where things got interesting. I asked the same clinical question two different ways:
Formal: “Is there radiographic evidence of cardiomegaly?”
Casual: “Does this show a big heart?”
Both questions mean the same thing. But the model’s internal features looked completely different.
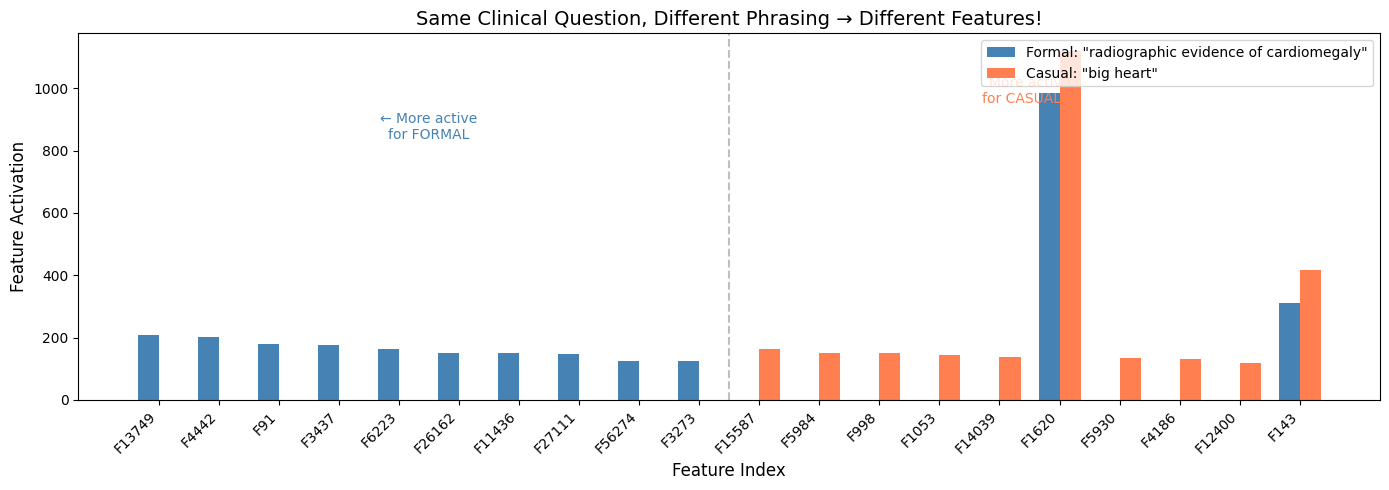
The cosine similarity between the feature vectors was 0.973. High, but not identical. And when I looked at the biggest differences:
Features more active for the formal phrasing:
Feature 13749: 207 vs 0
Feature 4442: 203 vs 0
Feature 91: 180 vs 0
Features more active for the casual phrasing:
Feature 15587: 163 vs 0
Feature 5984: 152 vs 0
Feature 998: 151 vs 0
Some features only fire for formal clinical language. Others only fire for casual phrasing. The model has learned distinct representations for how questions are asked, not just what they’re asking.
Experiment 3: Tracking Features Across a Phrasing Spectrum
In this experiment, I created a gradient of phrasings from very formal to very casual:
“Is there radiographic evidence of cardiac enlargement?”
“Does the imaging demonstrate cardiomegaly?”
“Is there cardiomegaly?”
“Is the heart enlarged?”
“Does this show a big heart?”
“Is the heart too big?”
“Big heart?”
Then I tracked two key features across this spectrum: Feature 13749 (most active for formal phrasing) and Feature 15587 (most active for casual phrasing).
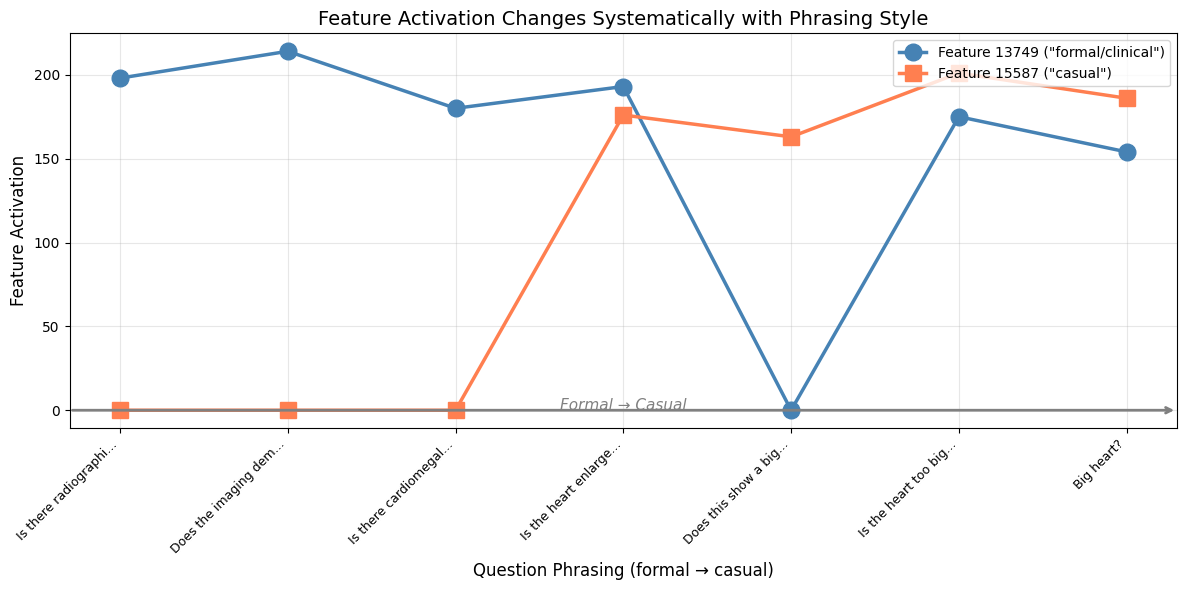
The pattern was clear. As the phrasing got more casual, Feature 13749 dropped and Feature 15587 rose. The crossover happened right around “Is the heart enlarged?” in the middle of the spectrum.This has real implications for medical VLM safety.A radiologist using formal clinical terminology might get a different answer than a patient asking the same question casually. The underlying clinical content is identical. But the model routes it through different internal circuits.
We already know medical VLMs can be sensitive to question phrasing. Now we can see why. The model isn’t just processing the semantic content of your question. It’s also encoding the style, the register, the formality. And those encodings influence what happens downstream.
For clinical deployment, this suggests we need:
Standardized prompting protocols
Robustness testing across phrasing variations
Feature-level monitoring for production systems
SAEs give us a lens into what’s happening inside these models. We can see which features drive a diagnosis, identify when features misfire, and understand why subtle changes in input lead to different outputs.The code is available as a Colab notebook. It runs on a free T4 GPU in about 5 minutes. If you’re working with medical VLMs, I’d encourage you to try it. See what features light up for your specific use cases. You might be surprised by what you find.
For more technical details, check out Anthropic’s original work on monosemanticity and the GemmaScope paper from Google. The SAEs are available on HuggingFace at google/gemma-scope-2-4b-it.
Also do check out the interactive demo in neuronpedia for Haiku Circuit tracer.